Section: Research Program
Research axis 2: Models for Molecular Evolution
Backwards models. The example of dating with transfers.
Here the goal is to compare extant genomes in order to propose plausible scenarios for their evolution. We constructed a multi-scale model of evolution, at the levels of species, chromosomes, genes and nucleotides. Reconstruction algorithms proposed a catalog of putative ancient transfers in different domains of life, as cyanobacteria, fungi, archaea. As transfers can only occur between contemporaneous species, they contain an information on the timing of life diversification. This allowed us to exploit a completely novel and abundant information for the history of biodiversity [81], [83], [82], [84], [45].
Forward models I. In silico experimental evolution.
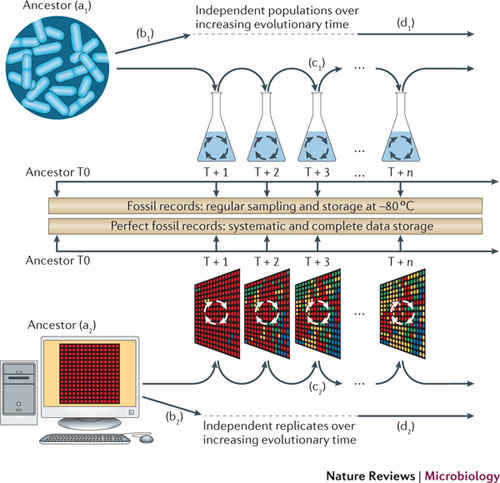
We have made several crucial advances in particular around the Aevol software that we have been developing for more than 10 years. This software consists in evolving in silico a population of digital genomes from an ancestral state and quantifying expected or unexpected systematic behaviors, in function of settled parameters (see Figure 1). We formalized the method [35] and implemented a tracking system that allows us to measure histories of genes [61]. We explored the potentiality of epigenetic inheritance through R-Aevol, an extension of Aevol implementing genetic regulation and epigenetic (cytoplasmic) inheritance [86], [87]. We also developed models with a new artificial chemistry in the context of the European project Evoevo (see below).
Forward models II. Theoretical investigations.
Simulations are made when models are too complicated to be handled analytically. However, it is sometimes worth simplifying the model in order to demonstrate a given property and have a proven behavior in function of perfectly identified parameters. We have obtained important results in this scope, mostly about the size of impact of selection on genome sizes [55]. Another set of theoretical results was obtained by the introduction of phenotypic noise in Fisher's Geometric Model (see below).
The Forward-Backward and the double blind procedure.
One objective of simulations or mathematical forward models is to explain the shape of some extant particular data. When one (or more) evolutionary mechanism is evoked to explain a given dataset, simulations can test if the invoked mechanism is indeed likely to produce the observed dataset. We investigated for example the causes of genome reduction in Prochlorococcus and compared them with genome reduction in endosymbiots [35], [36]. Another way we made the forward and backward models enrich each other is by using forward models to construct test instances for the backward methods. We have developed forward and backward models that are not scientifically linked, except by a common biological background [42], [41]. That led us to initiate the development of a new version of the Aevol software, “Aevol_ACGT”, that will include a four nucleotides genome decoded through a realistic genetic code [46], [63].
Current Objectives
We are improving and publicizing the dating method using gene transfers. We are leaders on this subject, published already in very good journals or labels, so we still see this subject as highly promising. A second important goal is producing benchmarks for the validation of evolutionary studies. This can be very useful for the evolutionary biology community, which lacks good validation standards. We provided some proofs of concept during the last years and will work towards a completely usable platform in the following years. Moreover we are pushing our in silico experimental model towards standard models of theoretical biology to test the robustness of some hypotheses to complex models. Finally we want to explore the possibility to co-construct biological knowledge with the general public through participative platforms. We will propose a protocol usable by the public in order to solve unknown parts of the tree of life, which size now calls for a participative project like encyclopedias or maps. This will demand new skills in computer science, participative sciences, and also will depend on consequent funding applications.